
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
ORIGINAL PAPERS / ARTICLES ORIGINAUX
MYOTONIC DYSTROPHY: CLINICAL AND MOLECULAR SPECTRUM IN KWA ZULU NATAL, SOUTH AFRICA
LA MYOTONIE DYSTROPHIQUE : ETUDE CLINIQUE ET MOLECULAIRE AU KWA ZULU NATAL, REPUBLIQUE SUD AFRICAINE
E-Mail Contact - MOTALA Ayesha :
mamot@sahcp.com
ABSTRACT Background and Purpose Method Results Conclusion Key words: Myotonic Dystrophy, Clinical Description. Molecular Diagnosis RESUME La myotonie dystrophique est la forme la plus commune de la dystrophie musculaire de l’adulte. Les type 1 et 2 de la myotonie dystrophique sont des désordres multisystemiques génétiquement dominant autosomique ; le type 1 avec une faiblesse principalement distale et le type – 2 avec une faiblesse principalement proximale. Le type 1 est provoqué par l’expression instable de trinucleotide de CTG. Le type – 2 est provoqué par un tétranucleotide, expansion de CCTG. Toutes les mutations peuvent être détectées en utilisant une combinaison des techniques de l’amplification en chaîne par But Méthode Résultats Conclusion Mots clés : Myotonie Dystrophique, clinique, diagnostic moléculaire, Sud Afrique INTRODUCTION Myotonic dystrophy is the commonest form of adult muscular dystrophy. Myotonic dystrophy type 1 and 2 (DM 1 and DM 2) are autosomal dominant inherited disorders with unusual multisystem clinical features characterized by myotonia, progressive muscle weakness and wasting, cataracts, hypogonadism, frontal balding, cardiac conduction defects and diabetes. Severity varies from asymptomatic to severely affected. (3) DM 1 is caused by the expansion of an unstable CTG trinucleotide repeat in the 3′ untranslated region of the myotonic dystrophy protein kinase gene on chromosome 19q13.3. (4, 6, 12) The postulated disease mechanism is thought to be the result of a reduction of the DMPK gene product, neighbour gene suppression (SIX5) and the processing of RNAs with longer CTG repeats. (1, 4) The CTG repeat is polymorphic in the general population. Healthy individuals have alleles with repeats lengths ranging between 5 and 35. A repeat length of between 35 and 49 is considered a “premutation” allele. Patients with DM 1 have expansions between 50 and several thousands. The size of the repeat is positively correlated with disease severity and inversely correlated with age of onset of symptoms. DM 1 is characterized by anticipation where affected individuals in succeeding generations have an earlier age of onset and more severe clinical course. (1, 4, 6, 12) Direct analysis of the CTG repeat expansion has a specificity and sensitivity of 100%. All DM1 mutations can be detected using a combination of the Southern Blot and Polymerase Chain reaction (PCR) techniques. (1, 3, 4, 6, 12) PCR is used to identify DM1 repeats between 5 and 200.Using specifically designed synthetic oligonucleotide primers based on the sequences flanking the triplet repeats, the unstable region can be easily amplified. If run on a 3, 5 % metaphor gel along with a size standard, the length of the repeat can be accurately determined. Repeats greater than 500 are not reliably amplified by PCR. Southern blot analysis of DNA digested with one of several restriction endonucleases (EcoR1, BamH1, Nco1, Bg1) is the procedure of choice for detection of CTG repeats greater than 200. Several Probes are available for hybridization. Using southern blot analysis, small-expanded alleles can be detected, which are seen to co-migrate with the normal allele during agarose gel electrophoresis after PCR is performed. This is usually difficult to resolve hence the use of Southern hybridization for these cases. The above are based on the new nomenclature and DNA testing guidelines for DM1 produced by the International MD Consortium. (2) DM 2 is linked to the long arm of chromosome 3q21. (13, 15) It is caused by a tetranucleotide, CCTG expansion in intron 1 of the zinc finger protein 9 (ZNF9) gene that interferes with processing of a variety of RNAs. (13, 15) DM 2 closely resembles DM 1 clinically. However, there are important differences. (5)
Prior to the discovery of the DM2 gene, 98% of patients with myotonic dystrophy demonstrated expanded CTG repeats on chromosome 19. (6,12,15) Numerous studies have been performed focusing on characterizing the molecular and clinical spectrum of DM 2 but the frequency with which it occurs in the general population or within the DM group is as yet uncertain. The exact incidence of DM2 will be easier to determine with increasing testing for the ZNF9 gene. (5) No data has been published regarding the frequency of both forms of DM in clinically affected DM individuals in KwaZulu – Natal (KZN), a province in South Africa. This study aims to characterize the clinical spectrum and molecular features of myotonic dystrophy patients in KwaZulu – Natal between 1989 and 2004. METHODS 1.1. Patient identification and assessment Patients included in this study were identified from the database of patients diagnosed with Myotonic Dystrophy at the Department of Neurology in KZN from 1989 to 2004. Consent for DNA analysis was obtained from each patient. Blood samples were collected in EDTA tubes. DNA extractions were performed using conventional procedures and PCR analysis of the patients DNA was performed at the Neuroscience Laboratory, University of Natal. Samples were sent to Department of Genetic Medicine, Sir Ganga Ram Hospital, Rajinder Nagar, New Delhi for analysis by PCR and southern hybridization. PCR ANALAYSIS IN UNIVERSITY OF NATAL NEUROSCIENCE LABORATORY Optimised PCR procedures using primers specific for DM1 were performed on all patient samples. A PCR assay was optimized using primers specific for DM2, and all patient’s negative for DM1 expansion were subjected to the DM2 PCR assay. PCR products were electrophoresed on a 2% agarose gel and analysed using the GelDoc software from Biorad. The PCR products were subsequently purified using the HighPure kit from Roche. The purified products were subjected to DNA sequence analysis using the 3100 Genetic Analyser.The results were analysed using the Biotools DNA sequence analysis software. Those patients’ samples found to not have a DM1 or DM2 duplication using the PCR assays were subjected to Southern hybridization using specifically designed probes. DNA analysis at Department of Genetic Medicine Sir Ganga Ram Hospital was done using the methods described in Current Protocols in Human Genetics (9) STATISTICAL ANALYSIS Data were captured in Microsoft excel and exported into SPSS version 11.5 (SPSS inc. Chicago, Illinois, USA) for statistical analysis. Simple one-way frequencies and bar charts were used to describe categorical variables. Quantitative variables were described using means and standard deviations. RESULTS Twenty five patients were identified from previous referrals to the neurology department. Eighty five percent of patients were of Indian decent and the remaining 15% were Caucasian. No African patients were identified in both the included and excluded patients. Sixty five percent were male and 35% female. In the majority (65%) the age of onset of clinical symptoms was below the age of 40 years. The average age of onset was 31.35. Twenty five percent had no family history of note. Of the remaining 75%, positive parental history was noted in 30%, 65% in siblings and 10% in offspring. No abnormalities were noted in the vital signs. One patient was on treatment for hypertension. Clinical characteristics are summarized in tables 1 and 2.Fifty five percent demonstrated a normal MMSE of 30. Twenty five percent demonstrated mild cognitive impairment of between 26-29 and 20% moderate impairment (20-25). Symptoms of hypersomnolence were found in 30% with 5% requiring treatment with methylphenidate. Basic blood investigations such as full blood count, renal function, liver function and other electrolytes were all normal. Elevated cholesterol was noted in 30%.One patient was on treatment for hypothyroidism. Three (15%) patients were being treated for diabetes mellitus type 2.Creatinine Kinase was mildly elevated in 50% .The highest level obtained was 899.The chest radiograph was normal in 80% with the remaining 20% having no available CXR. Electrocardiograph was performed in 75% of patients with 15 % demonstrating a left bundle branch block and 5 % a right bundle branch block. Nerve conduction studies were obtained in 60% of patient and were normal in 55%. The 1 patient demonstrated mild abnormalities with slightly decreased amplitude in median and peroneal nerves. Elelectromyography was performed in 85 % of patients with all demonstrating typical myotonic changes while 40% had evidence of myopathic findings as well. All patients were managed supportively with physiotherapy and occupational therapy. At the end of the study date 2 patients (10%) had demised and 1 (5%) was lost to follow up. The remaining patients were followed up either telephonically or with a recent visit to our clinic. Southern blotting performed at Sir Ganga Ram Hospital demonstrated expanded CTG repeats in all 20 samples. The PCR analysis was unable to demonstrate expanded alleles. Southern blot studies confirmed the presence of an expansion in all 20 samples for the CTG trinucleotide repeat found in myotonic dystrophy type 1. See figure 1. DISCUSSION All patients included in this study presented with predominantly distal weakness and a clinical assessment of type1 myotonic dystrophy was made. The molecular diagnosis confirmed the presence of expanded repeat for myotonic dystrophy type 1 supporting the clinical diagnosis. The study revealed that the majority of patients in KwaZulu – Natal who were affected by Myotonic Dystrophy were of Indian Descent. No patients of African origin were identified. A previous study by Lotz and Van Der Meyden (10, 11) in 1985 identified predominantly white patients in the northern Transvaal and no patients of African origin were documented. Goldman (7) in addition documented no South African Negroid patients with myotonic dystrophy. In addition Goldman (7) found that South African blacks have significantly fewer large repeat lengths than do white and Japanese populations and suggested that the occurrence of fewer large CTG repeats in the normal range may, in part, explain the absence of DM in southern African blacks. Goldman (8) further suggested that DM mutations in the Afrikaans population might have originated from a common initial founder who introduced one of the European ancestral mutations. The majority of patients demonstrated typical clinical features of myotonic dystrophy. EMG was diagnostic in those patients in whom they were performed. PCR did not demonstrate any expanded repeats. However the expansions were observed in all samples with southern blotting. While the southern blot expansions could not be quantified, they usually demonstrate expanded repeats when larger than 200.The negative PCR would therefore confirm that PCR can only be used to demonstrate expansions in patients with shorter repeats of between 5 – 200. All patients demonstrated clinical features of myotonic dystrophy type 1 and in view of the molecular findings this would suggest that patients with obvious clinical features probably possess larger repeat lengths, which would only be confirmed using southern blotting. PCR should therefore be used as a screening procedure for asymptomatic relatives of affected individuals to ascertain if they demonstrate the trinucleotide repeats and are at risk of becoming symptomatic at a later stage. Mildly symptomatic patients can also be screened with PCR, as they may possess shorter repeats. In conclusion, this study identified patients presenting with myotonic dystrophy in KwaZulu – Natal and demonstrated that Myotonic Dystrophy Type 1 remains the commonest clinical and molecular presentation. In addition it supported previous findings in which no South African of African decent was found to be affected by the disease. The molecular studies confirm that PCR is of limited value as only small repeats less than 200 can be demonstrated by this method. PCR may be important in asymptomatic individuals or for genetic counseling. Southern Blotting remains the gold standard in obtaining a molecular diagnosis. An important finding is that the clinical diagnosis was confirmed by molecular tests and this would suggest that clinical diagnosis is sufficient and molecular confirmation is not a requirement. The molecular testing would be of value in differentiating between DM1 and DM2 and in prenatal testing. Table 1. Frequency of clinical characteristics
Table 2. Frequency of distribution of wasting and weakness and extent of disability
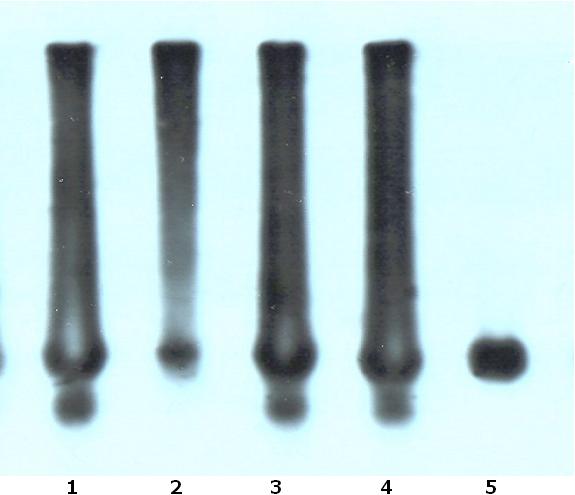 FIGURE 1 REFERENCES
|
© 2002-2018 African Journal of Neurological Sciences.
All rights reserved. Terms of use.
Tous droits réservés. Termes d'Utilisation.
ISSN: 1992-2647